We work at the spatial biology-technology interface to understand chromatin<–>synapse communication in the mammalian brain.
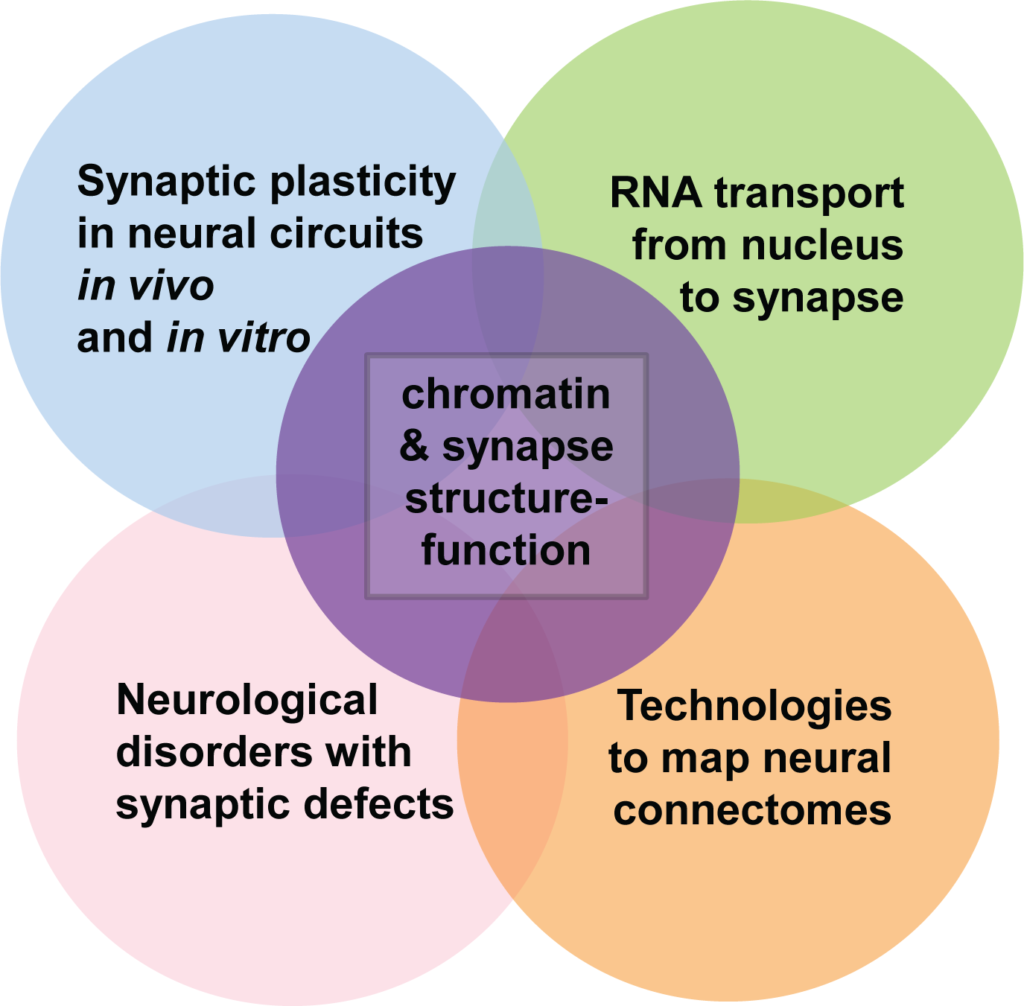
The Cremins Laboratory works at the spatial biology-technology interface to investigate the structure-function relationship of connections across the scales of chromatin, synapses, and circuits in the developing and diseased mammalian brain. Neurons exhibit elaborate structures at multiple scales, including diverse long-range axonal projections to form circuits, a range of complex dendritic spine geometries at the synapse, and the folding of the 10 nm chromatin fiber into compartments, topologically associating domains (TADs), and long-range looping interactions in the nucleus.
We have thus far focused on the nucleus to elucidate chromatin’s higher-order folding patterns at kilobase-resolution during neural lineage commitment, maturation, activity stimulation, and in neurological disorders. In mammals, the genetic sequence is >2 meters when stretched out end-to-end. It is folded into exquisite structures to fit into a nucleus roughly the size of a pin’s head. At the lab’s inception, it remained unclear how genomes are folded in a cell type-specific manner below the resolution of a Megabase, and whether and how higher-order folding could deterministically influence genome function. Our major contribution has been to provide early foundational insights into the genome’s structure-function relationship. We have elucidated how chromatin works through long-range mechanisms to govern the establishment of new gene expression, repeat instability, and replication origin firing patterns when mammalian cells transition states in the healthy and diseased brain.
Overall, by adding a spatial, third dimension to our understanding of chromatin’s regulatory mechanisms, the Cremins lab has uncovered foundational basic insights into the genome’s structure-function relationship in the mammalian brain and in neurological disorders. Looking forward, the lab will focus on mechanisms underlying chromatin-synapse communication and a fundamental mystery in neuroscience: how memory is stored over decades despite rapid turnover of proteins/RNAs. The long-term goal of the Cremins lab is to bridge chromatin folding, synaptic plasticity, and neurophysiology to elucidate how the genome’s structure-function relationship influences synaptic defects in intractable neurodevelopmental, neuropsychiatric, and neurodegenerative disorders.
-
William Kastor joins the lab as a new research technician. Welcome William!
-
Harshini Chandrashekar defends her thesis research and receives her PhD. Congratulations Dr. Chandrashekar!
-
Sarah Park joins us as our new laboratory manager. Welcome Sarah!
-
Ken Chandradoss presents a talk on his work at the Chromatin and Epigenetics Gordon Research conference in Newport, Rhode Island.
-
Katelyn Titus publishes a new paper on the link between RNA Polymerase II elongation and cell-type specific enhancer-promoter loops during human neuron differentiation:https://www.cell.com/cell-genomics/fulltext/S2666-979X(24)00200-3
-
Rohan Patel and Kenneth Pham submit their new manuscript on FISHnet – a tool for identifying chromatin domains in single-cell sequential Oligopaints imaging data. Congrats Rohan and Kenneth! https://www.biorxiv.org/content/10.1101/2024.06.18.599627v1
-
Rahul Sureka joins the lab from EMBL Rome as a Research Scientist. Rahul – we are thrilled to work with you on new projects. Welcome!
-
We are thrilled to share the news that our post-bacc alum James Sun will defend his PhD from NYU Grossman School of Medicine. James, we are filled with pride and joy to see all you have gone on to accomplish. Cannot wait to see what you achieve as you finish up your medical school training…
-
Delighted to share the news of renewal of a grant through the National Institute of Mental Health on 3D genome misfolding in human models of fragile X syndrome. Kenneth Pham, Thomas Malachowski, Chuanbin Su, Rohan Patel, Ravi Boya, and Ken Chandradoss will be working in part on projects related to this topic.
-
The Cremins lab is honored to announce a new partnership with the Cure Huntington’s Disease Initiative (CHDI). Han-Seul Ryu will be leading the initiative to study chromatin-mediated regulation of genome instability in single cells.
-
Constin Liu joins us from WUSTL as a new computational scientist in our SEEDs-BRIDGE post-bacc program. Welcome Constin!
-
Sarah Park joins the lab as our new lab manager. Welcome Sarah, we are all so happy to work with you!

Our work is supported by the New York Stem Cell Foundation, the Alfred P. Sloan Foundation, the National Science Foundation, the National Institute of Mental Health, the National Institute of Neural Disorders and Stroke, the National Science Foundation (NSF), NIH Common fund initiatives, the Friedreich’s Ataxia Research Alliance, the Chan Zuckerberg Initiative (CZI), the Cure Huntington’s Disease Initiative (CHDI), and the NIH 4D Nucleome Common Fund Initiative.
