We work at the spatial biology-technology interface to understand chromatin<–>synapse communication in the mammalian brain.
The Cremins lab aims to understand how chromatin works through long-range physical folding mechanisms to encode neuronal specification and long-term synaptic plasticity in healthy and diseased neural circuits. We pursue a multi-disciplinary approach integrating data across biological scales in the brain, including molecular Chromosome-Conformation-Capture sequencing technologies, single-cell imaging, optogenetics, genome engineering, induced pluripotent stem cell differentiation to neurons/organoids, and in vitro and in vivo electrophysiological measurements.
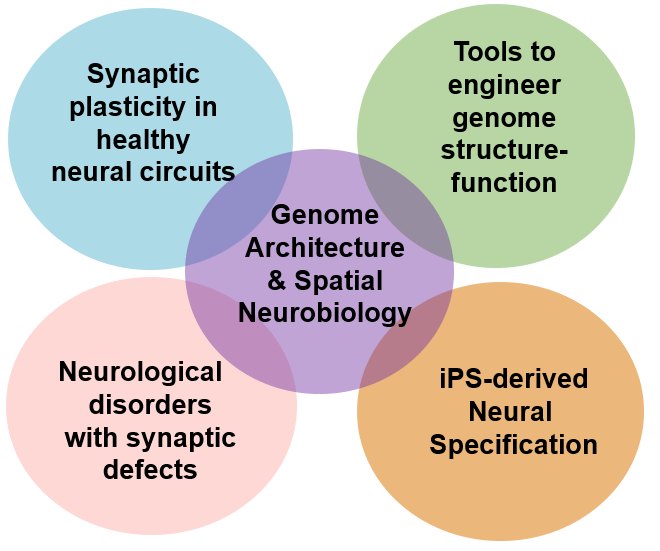
-
Warm congratulations to Harshini, Zoltan, Heesun, Avi, Ali, and Han-Seul for posting their new BioRxiv preprint uncovering a role for dynamic multi-loops in gene expression dysregulation in post-mitotic human neurons homozygously-engineered with familial Alzheimer’s disease mutations. https://www.biorxiv.org/content/10.1101/2024.02.27.582395v1
-
Congratulations to Dr. Peibo Xu – one of his first-author PhD papers entitled ‘High-throughput mapping of single-neuron projection and molecular features by retrograde barcoded labeling‘ is now published in eLife. Congratulations Peibo!https://pubmed.ncbi.nlm.nih.gov/38390967/
-
MSTP student Han-Seul Ryu passes her proposal and becomes a PhD candidate. Congratulations Han-Seul!
-
Huge congratulations to MSTP student Kenneth Pham on receiving his first NIH Notice of Award for his F30 fellowship grant. Great job Kenneth!
-
Edda Schulz writes a commentary on our team’s fragile x paper in Molecular Cell. https://www.sciencedirect.com/science/article/pii/S1097276524000017?dgcid=author
-
NIMH writes a news article on our Fragile X Syndrome work: https://www.nimh.nih.gov/news/science-news/2024/researchers-expand-understanding-of-genetic-mechanisms-underlying-fragile-x-syndrome
-
Kenneth Pham receives a fundable score on his NIH NRSA F30 proposal
-
Warm congratulations to our Ken Chandradoss on his marriage!
-
Thomas, Ravi, Ken, and Linda publish their paper ‘Spatially coordinated heterochromatinization of long synaptic genes in fragile X syndrome’ – warmest congratulations to the lead authors and all members of Team FXS for their extraordinary resilience, rigor, and dedication to seeing this through to the finish line. https://doi.org/10.1016/j.cell.2023.11.019
-
Harshini Chandrashekar, Chuanbin Su, Katelyn Titus, and Rohan Patel present posters on their work at the 4DN annual meeting.
-
Harshini Chandrashekar gives a talk at the 2023 4DN annual meeting.
-
Lab heads to Boston for the 2023 4DN annual meeting. Make way for ducklings!
Our work is supported by the New York Stem Cell Foundation, the Alfred P. Sloan Foundation, the National Science Foundation, the National Institute of Mental Health, the National Institute of Neural Disorders and Stroke, NIH Common fund initiatives, the Friedreich’s Ataxia Research Alliance, the Chan Zuckerberg Initiative, and the NIH 4D Nucleome Common Fund Initiative.

